Dasantila Golemi-Kotra, York University, Canada
More and more countries around the world are struggling with a new wave of infections, with an alarming increase in COVID-19 cases. Mostly these new surges are attributed to variants of SARS-CoV-2.
In lay terms, a variant refers to a virus genetically distinct from its original strain. The emergence of variants in the microbial world is nothing new, just think of “superbugs” such as MRSA (methicillin-resistant Staphylococcus aureus), which is resistant to almost all the existing antibiotics.
How did the U.K., South Africa and Brazil variants emerge?
Genetic transformation of a virus occurs by mutation. Mutations emerge naturally in the microbial world. They are mistakes in the genetic code caused by copying processes in the cell. These mistakes are exploited by the virus to survive and establish itself, especially under adverse conditions.
Curiously, viruses mutate at a much higher rate than other micro-organisms. Often mistakes in the genetic code come and go without leaving any traces, but in a few instances, they are selected when they offer the microbe a growth (or infection) advantage.
Read more: Drug-resistant superbugs: A global threat intensified by the fight against coronavirus
In the case of SARS-CoV-2, several mutations have been selected, meaning they are propagated to the next generation of viruses. They are selected for efficient access to host cells — fewer viruses are needed to infect the host — and effective evasion of the neutralizing antibodies of the immune system, meaning they are able to dodge immune response so they can circulate longer in the host, providing more opportunity to infect other cells.
However, human intervention — such as the use of certain treatments like convalescent plasma or monoclonal treatment — can drive virus evolution in much the same way that antibiotic use drives the evolution of bacterial superbugs.
How seriously should we take SARS-CoV-2 variants?
The World Health Organization (WHO) categorizes virus variants into two different groups: variant of concern (VOC) and variant of interest (VOI). VOCs represent those variants that are linked to the rise of new infection waves in many countries, including the recent surges in Canada and United States.
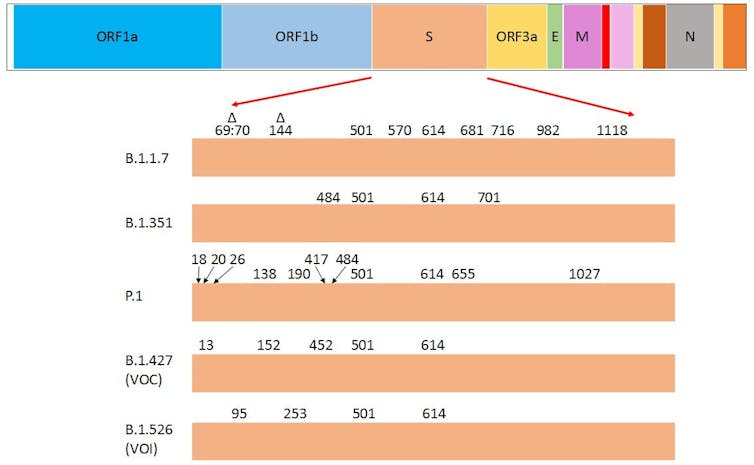
Here in Canada, the B.1.1.7 variant, which emerged in the United Kingdom in September 2020, is becoming the dominant variant.
The other VOCs — such as B.1.351, first identified in South Africa, and P.1, first identified in Brazil — are being identified more often and have been responsible for several outbreaks in Canada.
There is evidence, some of which has not yet been peer reviewed, that VOCs are associated with higher virulence than the coronavirus that originated in Wuhan, China: higher transmissibility, possible higher disease severity and, in the case of B.1.351, an increased ability to evade neutralizing antibodies.
These attributes of VOCs have translated into higher hospitalizations of younger people and an increase in deaths across all age groups in Canada.
VOIs are on the radars of public health agencies for their impact on virus transmission, the severity of disease and vaccine effectiveness.
How does mutation change how a virus works?
The genetic code of a virus provides instructions to make its proteins: strings of amino acids in defined sequences. Mutation can lead to amino acid substitution or deletion in the protein. The attention of the scientific community is focused on the amino acid substitutions that affect the SARS-CoV-2 spike protein, the protein that gives the virus its crown-like shape.
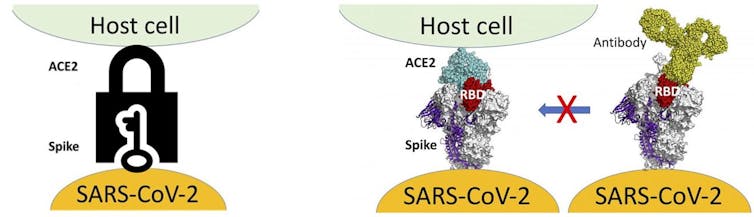
The spike protein is the key that provides access to human cells via the human ACE-2 protein (the lock), and as such is the target of the currently approved COVID-19 vaccines. Research that has yet to be peer reviewed shows two ways in which changes to the amino acid sequence of the spike protein may affect its interaction with human cells:
- They can enhance the spike protein’s interaction with ACE-2, providing efficient access to host cells.
- They can decrease the spike protein’s interaction with neutralizing antibodies, helping it evade an immune system response long enough to infect other cells.
Proteins can be thought of as microscopic Lego structures, with amino acids behaving as Lego pieces and held together by a single thread. However, protein structures are much more flexible than Lego structures (think of Lego pieces made of Jell-O), with the amino acids able to form transient bonds with other amino acids close to them as needed for structural stability and recognizing other structures.
The spike protein’s structural flexibility allows it to sample the space inside the lock (ACE-2) to enable recognition of the key, but also to find the optimum key-shape for that lock. It is the latter function that is optimized through mutation: the best key-shape will open the lock faster and more easily.
Could mutation lead to a super-variant that can evade all vaccines?
Many VOCs and VOIs have been identified around the world, and more variants get reported daily (about one million variants have been recorded to date). Should we be afraid of the emergence of a super-variant, which is highly virulent and can overpower all current vaccines and any others in the future?
The three current VOCs that have taken hold in many countries carry several amino acid substitutions in their spike proteins. Because of the spike protein’s crucial role in entering cells, all of the COVID-19 vaccines currently available work by targeting the spike protein.
Peculiarly, a few of the amino acid substitutions in the spike protein are common to the three current VOCs and are considered to drive their dominance over other variants. Research that has yet to be peer reviewed shows the substitutions N501Y (asparagine to tyrosine) and D614G (aspartate to glycine) are common to all three, while E484K (glutamate to lysine) is common to B.1.351 and P.1.
The E484K substitution is believed to be responsible for antibody evasion, and N501Y and D614G are thought to drive the higher transmissibility of these variants. (The CDC website offers more information on the attributes of the variants.)
It is curious that the common amino acid substitutions identified among VOCs emerged independently and in different parts of the globe. There are, in fact, 20 different amino acids provided by the host cell, all of which have an equal opportunity to substitute an amino acid in the protein through mutation. Yet, remarkably, these three VOCs evolved to acquire some of the same amino acid substitutions!
This phenomenon is referred to in biology as convergence evolution: when the same feature evolves independently. This means that the selected amino acids offer a unique property that makes the virus “fitter.”
The spike protein has an important function: it has to unlock access to the host cell, but it is also the target of neutralizing antibodies, which the virus has to prevent from latching onto the protein in order to evade the immune system. Both these functions rely on the same part of the protein: the receptor binding domain (RBD).
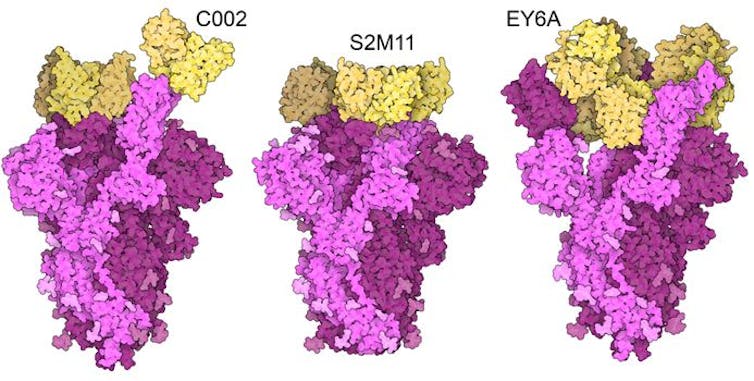
Enhancement of one of these two functions can diminish the other function. That means a compromise has to be reached. The fact that the same substitutions arose independently in different variants is an indication that the spike proteins in these variants have been optimized, and may not gain any further advantages.
Based on this, emergence of a super-variant seems unlikely, because these two functions — unlocking the host cell and evading the immune system — will always be in competition with each other, so neither will be able to achieve perfect efficiency.
However, never underestimate the force of evolution when there is a fertile playing field. We must adhere to public health measures, such as social distancing and masks, to mitigate the spread of the virus and limit the number of hosts that the virus can infect, for as long as most of the population remains unvaccinated.
Dasantila Golemi-Kotra, Professor, Biology, York University, Canada
This article is republished from The Conversation under a Creative Commons license. Read the original article.

