
Fathima Afsal, McGill University and Dominic Frigon, McGill University
From sore throats to fevers and life-threatening infections, most people have periodically used antibiotics. Recent reports show that the global COVID-19 pandemic has increased the use of antibiotics.
Some microbes may be naturally resistant to certain antimicrobials. In other cases, improper use of antimicrobials promotes the modification of microbial genes (genes are units of DNA), making the microbes resistant to antimicrobials. The man behind the discovery of antibiotics, Alexander Fleming, gave an anticipatory warning on this topic in his 1945 Nobel lecture:
“Mr. X. has a sore throat. He buys some penicillin and gives himself, not enough to kill the streptococci (bacteria known to cause sore throats and tonsils) but enough to educate them to resist penicillin. He then infects his wife. Mrs. X gets pneumonia and is treated with penicillin. As the streptococci are now resistant to penicillin the treatment fails. Mrs. X dies. Who is primarily responsible for Mrs. X’s death?”
This situation is termed antimicrobial resistance and the genes that confer resistance are referred to as antimicrobial resistance genes (ARGs).
Antimicrobial resistance genes
Globally, health problems due to antimicrobial resistance have been rising. One of the major reasons is the mobility of ARGs. The resistance genes can be transferred from one generation of microbes to the next through reproduction, and can also be transferred by horizontal gene transfer (non-sexual transfer of genetic material between different microbes).
This implies that regardless of their origin, antimicrobial resistance genes can be transmitted from environmental sources to the human gut, and vice-versa.
As environmental engineers, our research at McGill University focuses on developing tools for monitoring the movement of antimicrobial resistance genes in wastewaters, and understanding the impact of ARG transfer to human gut microbes. Apart from the person-to-person transmission of resistant microbes described by Fleming, there are several other ways these microbes can enter the human body.
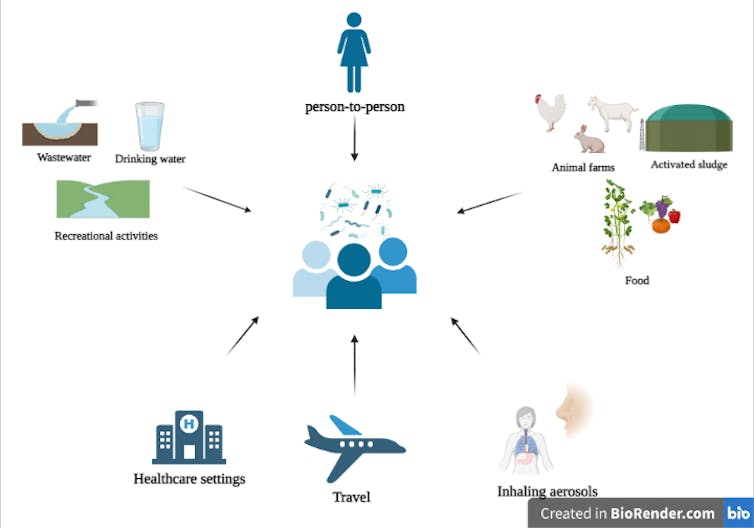
• Wastewater
Wastewater is a sink for human activities and contains resistance genes from human feces, farms, households and hospitals. Treated wastewaters often are released into surface waters such as rivers, which are used for recreational activities like sports, fishing and swimming. A cross-sectional study revealed that the gut of surfers is three to four times more likely to be colonized by multi-drug resistant microbes compared to the gut of non-surfers.
In addition, potable water is obtained from treated surface or groundwater. Research has shown that drinking water biofilms (aggregation of microbes living on surfaces) can transfer antimicrobial resistance genes to the mouse gut, indicating that they might also be transferred to the human gut.
• Food
To increase animal productivity, antimicrobials are often applied on farms, and this can lead to the selection and development of resistance genes. The application of animal manure and sludge (byproduct of wastewater treatment) in agriculture can mediate the transfer of antimicrobial resistance genes to food crops. Several ARGs have been found to travel from soil to edible parts of plants such as tomato, lettuce and broad bean crops.
• Air
Inhalable antimicrobial resistance genes pose an increasingly growing silent health threat. Recent reports from hospital samples have revealed that the daily human exposure to resistant bacteria contained in aerosols is 10 times higher than those present in drinking water.
• Health-care facilities
Health-care facilities are one of the most prominent locations for acquiring multi-drug resistant infections due to the presence of opportunistic pathogens that infect immunologically compromised patients. Presence of antimicrobials further aid in the development and selection of resistant microbes, which then can be transferred from one patient to others by interpersonal contact or via common surfaces such as door handles, bed rails or lockers.
• Travel
International travellers who travelled to developing countries have been found to acquire multi-drug resistant bacteria. They may have acquired these microbes via contaminated food and water, due to poor hygiene practices or because of differential antimicrobial usage policies across countries.
A One Health approach
It is evident that antimicrobial resistant microbes surround us, and it is normal to feel concerned. But what is even more important to recognize are the tiny steps we all can still adopt. This includes ensuring patients are treated with the appropriate antibiotic in the right dose.
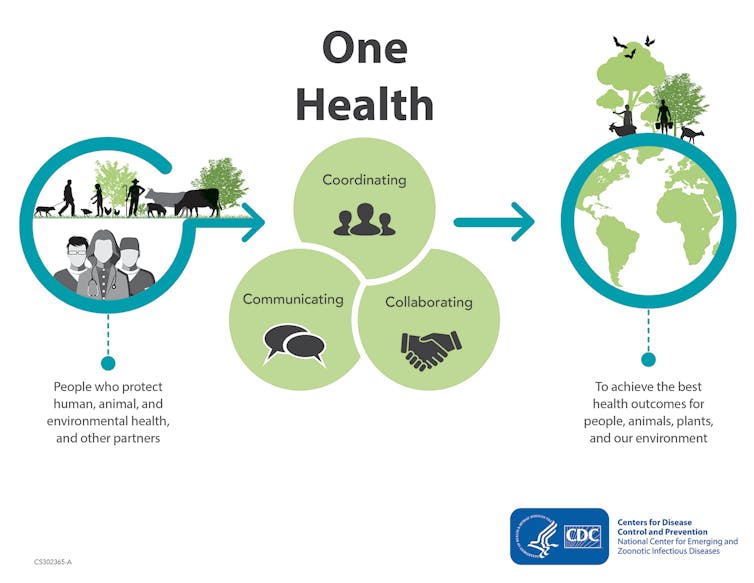
On a global level, understanding the transmission routes for resistant microbes is a part of a One Health approach. The holistic concept of One Health acknowledges that the health of humans is interconnected to the health of animals and our surrounding environment. This enables bringing together resources and policies to monitor and tackle antimicrobial resistance.
Indeed, resistant microbes are running a marathon with antimicrobials, and we need to take measures to slow down their pace. In the long run, this would ensure that the antimicrobials would continue to support human survival.![]()
Fathima Afsal, PhD student in Environmental Engineering, McGill University and Dominic Frigon, Associate Professor of environmental engineering, Civil Engineering Department, McGill University
This article is republished from The Conversation under a Creative Commons license. Read the original article.

