
Gabriel Wainer, Carleton University; Cristina Ruiz Martin, Carleton University, and Hoda Khalil, Carleton University
Pandemics are not new. We have historical records on the effects of pandemics dating from as early as 3000 BC. Between 1348 and 1350, the Black Death killed a quarter of the population in Europe. A century later, European diseases killed large numbers of Indigenous people in what is now known as Canada and the rest of the Americas.
The 1918 Spanish flu caused around 50 million deaths worldwide. Since then, we have suffered deadly outbreaks of smallpox, pertussis, Ebola, SARS, Avian flu and many others.
Although natural observations and social experiments allow us to study past diseases and their patterns, this is not enough to prevent future crises. To efficiently predict the spread of a disease, there is a clear and urgent need for new tools and methodologies that can easily assimilate possible factors, such as weather patterns and human behaviour.
If we could provide clear predictions on which social behaviours would affect the spread of a certain disease, policy-makers would be in a better position to develop improved pandemic response plans. Researchers are building new prediction models and running simulations to study how to deal with the current outbreak.
Our research group has defined new methods and distributed simulation techniques to study different aspects of the spread of disease. Our research also studied diffusion processes based on social interactions, like those happening during epidemics.
New viral outbreaks
We are currently dealing with a pandemic caused by SARS-CoV-2, a new coronavirus. The latest figures for confirmed cases and deaths point out that we were not prepared to deal with contagion rate and severity of this virus. Decision makers cannot wait until real data from new diseases are available to start planning. Recent research confirms that these outbreaks will keep on happening, so what can we do to be prepared for the next (inevitable) outbreak?
Simulating futures
Modelling and simulation methodologies can pave the way to improve future response plans. A model is a representation of the real world using mathematical equations. A simulation executes models using a computer to reproduce multiple cases over a period of time. Modelling and simulation, augmented with data from previous social events and diseases, can be effective prediction tools.
A recent popular article in the Washington Post shows a simulation about the effects of isolation in the spread of COVID-19. This is a simple model useful for education and understanding the consequences of policies, but models used for prediction are much more complex.
There are numerous methodologies for modelling and simulation. Methods that use sound mathematical theories produce higher quality and more efficient predictions.
Epidemiologists and other experts build models, and then work together with computer simulation experts to test their hypotheses. These models need to be easy to understand and update. Experts need to modify models easily to include factors that may change the evolution of an outbreak (for instance, environmental and social conditions).
The simulations should run quickly and efficiently, as we need to conduct a large number of different simulation experiments to obtain meaningful results. These results can help decision makers to make better choices and affect policy accordingly.
For epidemics, one can start with historical data from previous outbreaks, but as soon as new data is obtained, we must update the models and execute a large number of new simulations efficiently.
Predicting outbreaks
Our research group designed mathematical modelling and simulation tools for the visualization and analysis of complex social systems where the overall behaviour should study the individuals as well as their interactions. The methods improve the quality of the models and the speed of the simulations by separating how individuals and their interactions are modelled.
Take this simulation of H5N1, a form of avian flu and a highly contagious viral disease that spreads among poultry and other species. Our methods provide a complete solution to model, simulate and visualize the disease, and a mechanism to include diverse factors that may contribute to the outbreak outcomes.
One example of those factors include
. Other factors are the ability of a community to combat the infection, the probability of becoming infected based on the distance to an infected individual or the effects of vaccination. Our methods allow the modelling of complex behaviour related to the passage of time easily, including adjusting for weather, variation in incubation periods of viruses, etc.Expanding communication
Another important aspect for modelling and simulation during a pandemic is global collaboration between experts. Having models and simulations available globally permits sharing resources and working collaboratively. By making tools accessible online, researchers can execute the simulations on remote powerful computers when available and then visualize the results in personal devices to facilitate analysis and experimentation.
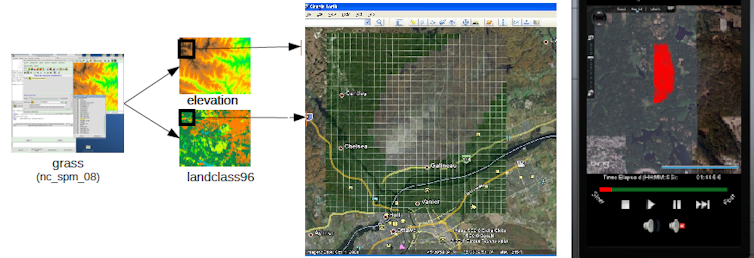
A mix of web services and geographical information systems help these share models with other people and study the results of simulations. Uploading models onto the cloud provides access through a variety of internet-connected devices (e.g. mobile phones, tablets, laptops).
Policy makers can study simulation results without installing complex software and epidemiologists can use and share resources. Results can be retrieved by modeller’s software engineers, scientists and policy makers for further analysis and improvements.
Simulation models can provide key insights for future pandemic response plans. These predictions will be a powerful tool in the attempt to inform the public and reduce casualties.
Gabriel Wainer, Professor, Systems and Computer Engineering, Carleton University; Cristina Ruiz Martin, Postdoctoral fellow, Systems and Computer Engineering, Carleton University, and Hoda Khalil, Postdoctoral fellow, Engineering, Carleton University
This article is republished from The Conversation under a Creative Commons license. Read the original article.

